Long read services
Long-read sequencing is an advanced next-generation sequencing (NGS) technology that generates reads significantly longer than those produced by traditional short-read sequencing methods. These longer reads, which can range from several kilobases (kb) to megabases (Mb) in length, offer distinct advantages in genome assembly, structural variant detection, and the analysis of complex genomic regions.
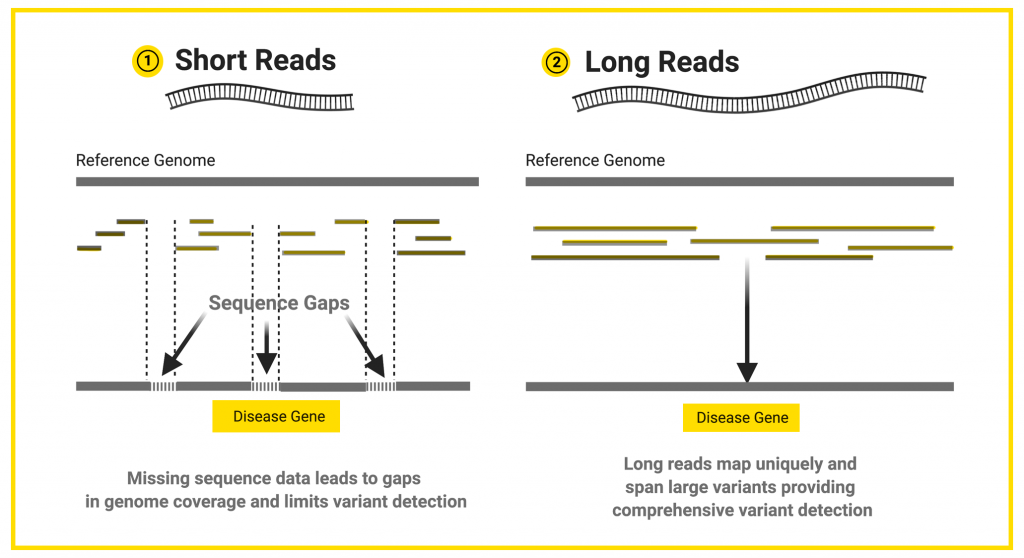
Credit: Sarah Sharman
Here's an overview of long-read sequencing:
Key Technologies:
-
PacBio Single-Molecule Real-Time (SMRT) Sequencing:
- Technology: Uses zero-mode waveguides (ZMWs) to sequence single DNA molecules in real-time. DNA polymerase incorporates fluorescently labeled nucleotides, and the fluorescence is recorded as the polymerase synthesizes the complementary strand.
- Read Length: Typically 10-25 kb, with some reads extending beyond 100 kb.
- Applications: De novo genome assembly, isoform sequencing (Iso-Seq), structural variant detection, epigenetic analysis (e.g., DNA methylation), and resolving complex regions like repetitive sequences.
- GCTU Service: Full length 16S, WGS, Shotgun Metagenomics, Iso-Seq, MAS-Seq, Kinnex workflows, Sequencing Only
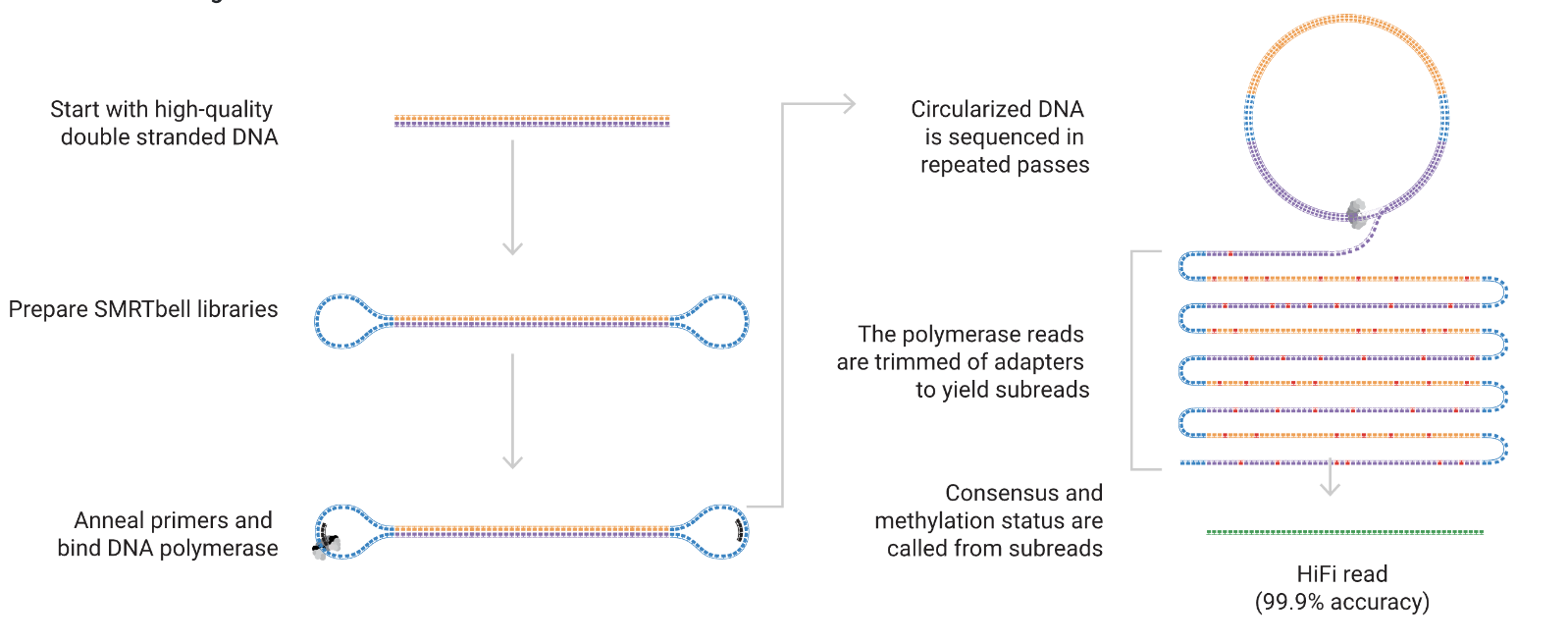

Credit: PacBio
2. Oxford Nanopore Sequencing:
-
- Technology: Passes single-stranded DNA or RNA molecules through nanopores embedded in a membrane. As the molecules move through the nanopore, changes in the ionic current are measured and interpreted as nucleotide sequences.
- Read Length: Capable of producing ultra-long reads, often exceeding 100 kb, and can theoretically sequence entire chromosomes in a single read.
- Applications: Whole-genome sequencing, real-time sequencing (e.g., pathogen identification), analysis of long-range haplotype information, detection of structural variants, and RNA sequencing.
- GCTU Service: cDNA-PCR, Custom, Sequencing Only
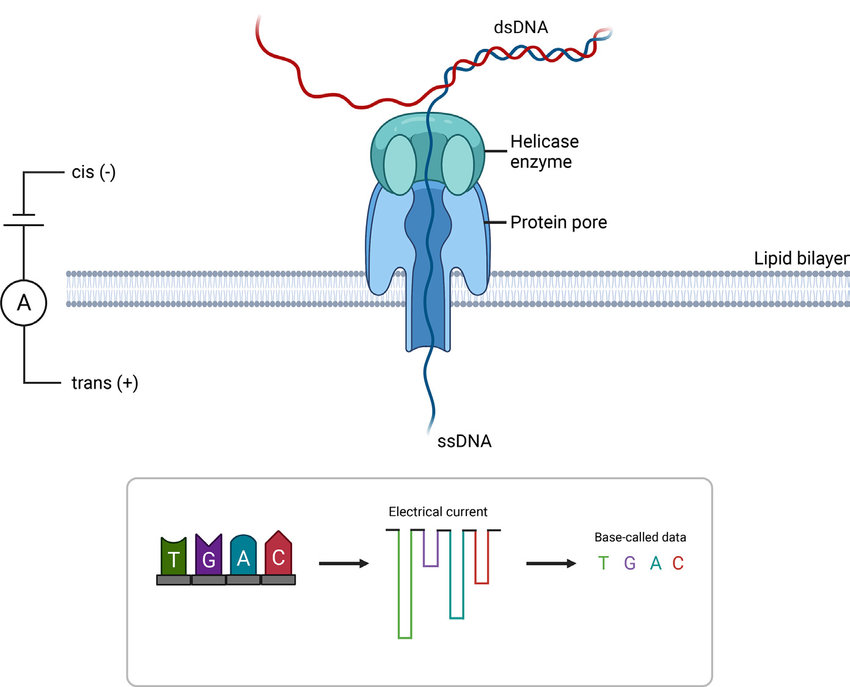
Credit: Beckett et al 2021
Advantages of Long-Read Sequencing:
-
Improved Genome Assembly:
- De Novo Assembly: Long reads simplify the assembly of genomes by spanning repetitive regions and resolving complex structural variants, leading to more complete and contiguous assemblies.
- Hybrid Assemblies: Long reads can be combined with short reads to enhance assembly accuracy and resolve regions that short reads alone cannot.
-
Structural Variant Detection:
- Large Variants: Long reads can detect large insertions, deletions, inversions, and translocations that are often missed by short-read sequencing.
- Complex Rearrangements: The ability to span entire structural variants allows for precise characterisation of complex genomic rearrangements.
-
Resolution of Repetitive Regions:
- Tandem Repeats: Long reads can sequence through repetitive regions, such as tandem repeats and transposable elements, which are challenging to resolve with short reads.
- Telomeres and Centromeres: Long reads are effective in sequencing through these highly repetitive regions, leading to more complete assemblies of these parts of the genome.
-
Haplotype Phasing:
- Phasing: Long reads enable the differentiation of maternal and paternal alleles across large regions of the genome, allowing for accurate haplotype phasing and analysis of genetic variation.
-
Isoform Sequencing (Iso-Seq):
- Full-Length Transcripts: Long-read sequencing allows for the sequencing of full-length RNA molecules without the need for assembly, providing a comprehensive view of transcript isoforms and alternative splicing events.
Challenges and Limitations:
-
Error Rates:
- Higher Error Rates: Some Long-read sequencing technologies can have higher raw error rates compared to short-read sequencing, though advances in technologies, bioinformatics and consensus algorithms have improved accuracy.
-
Cost:
- Expense: Long-read sequencing can be more expensive per base compared to short-read methods, though costs are decreasing as the technologies mature.
-
Throughput:
- Lower Throughput: Long-read sequencing generally produces fewer reads compared to short-read technologies, which can limit its utility for applications requiring very high coverage.
Applications:
- De Novo Genome Sequencing: Long-read sequencing is ideal for sequencing novel genomes, especially for organisms with complex genomes, where it provides more complete and accurate assemblies.
- Clinical Diagnostics: Used for identifying structural variants and large rearrangements in genetic disorders that are not easily detected by other methods.
- Cancer Genomics: Helps in identifying large-scale mutations and rearrangements in cancer genomes, contributing to more precise diagnostics and personalised medicine.
- Microbial Genomics: Provides complete assemblies of microbial genomes, including plasmids and other mobile elements, crucial for understanding pathogenicity and antibiotic resistance. Enables sequencing of the entire 16S rRNA gene, which is commonly used for microbial identification and taxonomy.
Long-read sequencing is a powerful tool that overcomes many of the limitations of short-read sequencing, particularly in genome assembly and the detection of complex genomic features. While it is still evolving and can be more costly and error-prone than short-read methods, its unique capabilities make it invaluable for certain applications, especially where accuracy and completeness of genomic information are critical. As technology continues to advance, the adoption of long-read sequencing is expected to grow, further expanding its applications in genomics and beyond.
In addition to off the shelf and custom services, we also offer 'Sequencing Only' service for user-prepped libraries on our long read instruments. Please see our Services page for more information.